Topic 7 signals the start of the higher-level curriculum, and the return to molecular biology. This topic takes a deeper dive into DNA structure, replication, transcription, and translation. The first part of this chapter was covered in topic 2, but I will explain it with more detail in this topic. I found this topic to be rather challenging, but also very interesting. The key to understanding DNA replication and translation is to make sense of the direction the different strands go in, and what direction the enzymes go in. Otherwise, it is quite a dense chapter, as a lot of information is squeezed into just a few pages.
Points for revision:
- Translation and protein synthesis
DNA and RNA structure
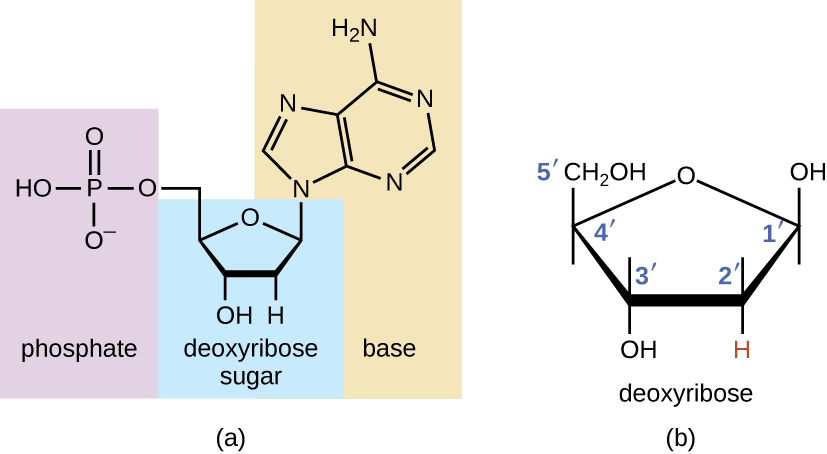
Deoxyribonucleic acid (DNA) is made of two key components, the sugar phosphate backbone and the nitrogenous bases. Ribose is a 5-carbon monosaccharide with one oxygen in the ring-like structure. Ribose is usually depicted with the oxygen pointing up, and the carbon to the right of the oxygen is called the 1’ (one prime) carbon. The numbering then follows a clockwise rotation ending with the 5’ carbon. In DNA, the 1’ carbon is bonded to a nitrogenous base, and the 5’ carbon bonded to a phosphate. This is why it is called a sugar phosphate backbone. In DNA, the 5’ end is always bonded to a phosphate, but the last 3’ end is not bonded to a phosphate group. The reason why will be explained further down the page in DNA replication. Since DNA has a double helix structure, and the strands run antiparallel to each other, one strand goes in a 5’ to 3’ direction, and the other strand goes in a 3’ to 5’ direction.
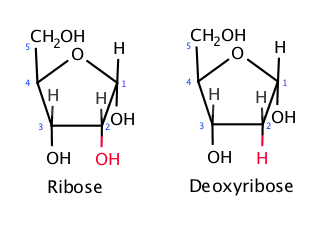
The other key component are the nitrogenous bases. It is in the nitrogenous bases where the code of life is stored. In DNA there are 4 bases, adenine (A), thymine(T), guanine(G) and cytosine(C). These bases always bond in complimentary base pairs: A with T, and C with G. This is because A and T can have two hydrogen bonds, while C and G can have 3. A and G are purines, meaning that they have a double ring structure, while C and T and pyrimidines, meaning that they have a single ring structure. A purine is always bonded to a pyrimidine.
A nucleotide is a ribose with the 5’ end bonded to a phosphate group, and the 1’ group bonded to a nitrogenous base. DNA is a long chain of nucleotides.
RNA (ribonucleic acid) is quite similar to DNA, but they have a few distinct differences. Firstly, RNA is single stranded, meaning that it only has one strand. Secondly, RNA does not have thymine, instead it has uracil (U) bonded with adenine. And lastly, the ribose in RNA has one oxygen more. The removed oxygen in DNA is why it is called “deoxy” ribonucleic acid.

DNA sequences
Different segments of the DNA sequence have different purposes. As you may recall, DNA in cells is in the form of chromatin when it is not in mitosis. Chromatin is DNA that is coiled around 8 proteins called histones. The resulting complex is called a nucleosome. However, the tight wrapping inhibits transcription enzymes to read the DNA around the nucleosome. This means that the DNA around nucleosomes cannot be transcribed. This is an example of non-coding DNA.
Other examples of non-coding DNA are structural DNA and highly repetitive sequences. Structural DNA appears near the centromeres and telomeres, and the highly repetitive sequences seem to appear randomly throughout the genome. It is still quite unclear what functions non-coding DNA serve. However, as you will see later, some non-coding DNA is important in regulating DNA transcription.
Protein coding DNA does as the name suggests, code for proteins. The DNA is first transcribed over to mRNA, then processed and transported out of the nuclear envelope to be attached to a ribosome. The ribosome then functions as a construction site, reading the code and attaching tRNA with amino acids, thus forming a polypeptide chain. This process is called protein synthesis and involves transcription and translation, which we will come back to later.
DNA replication
DNA replication occurs in the S phase of interphase, where the cell prepares for mitosis. It is the process where the cell duplicates its genetic material so that it can form a new identical cell. The process of replication involves a series of enzymes each with its own function. The first enzyme is the helicase. Helicase splits open the two intertwined DNA strands, thus separating them into two strands that run in opposite directions. The strand that runs in the 3’ to 5’ direction is called the leading strand, while the strand that runs in a 5’ to 3’ direction is called the lagging strand. The leading strand is quite easy to deal with, while the lagging strand is a little more annoying. The place where the helicase split open the DNA is called the replication bubble and it runs bilateral, which means that it goes in both directions. This speeds up the replication process. Additionally, multiple replication bubbles present simultaneously further speeds up the process.
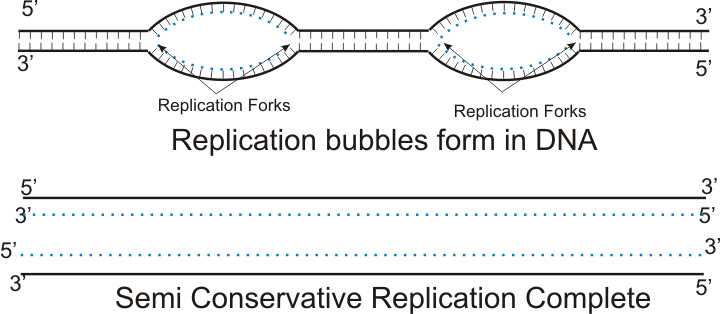
As previously mentioned, elongation (adding nucleotides to the new strand) of the leading strand is much easier to deal with than the lagging strand. This is because the enzyme that adds nucleotides to the new strand can only do it in a 5’ to 3’ direction. Since the leading strand runs in a 3’ to 5’ direction, the enzyme can just zip along in a continuous line. The enzyme that reads and adds nucleotides by complimentary base pairing is called DNA polymerase III. However, DNA polymerase III cannot start the process by itself, it needs a primer. An enzyme called primase therefore first adds a short RNA primer to the DNA strand, before the DNA polymerase takes over adding the rest of the nucleotides. Lastly, an enzyme called DNA polymerase I removes the RNA primer and replaces it with DNA nucleotides instead.
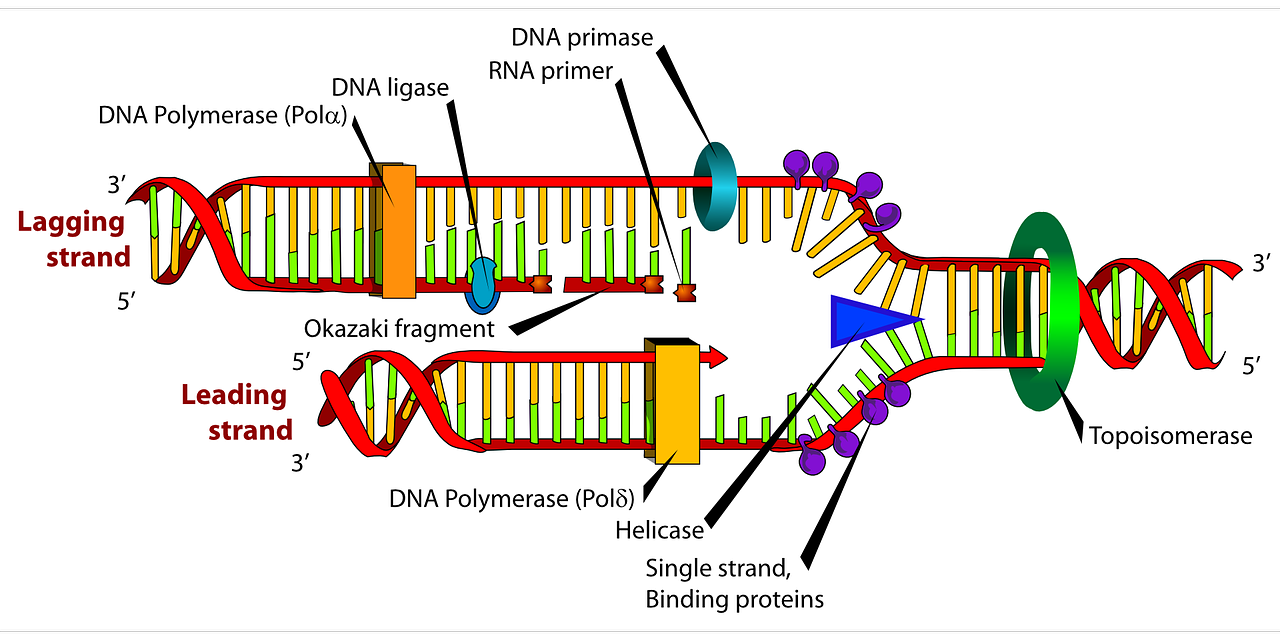
Now we need to deal with the lagging strand. Since DNA polymerase III can only add nucleotides in a 5’ to 3’ direction, it has to go backwards on the lagging strand since it also runs in a 5’ to 3’ direction. This is problematic since as the replication fork moves, more and more DNA become exposed. The DNA polymerase III therefore has to add nucleotides in chunks. As always, a primase adds an RNA primer which the DNA polymerase III starts elongating from. However, the primase then has to go back to the newly exposed DNA since the replication fork has moved and add a new primer. The DNA polymerase III then also has to move back and add a new chunk of DNA. Elongation of the lagging strand therefore happens in chunks of a primer and DNA nucleotides called Okazaki fragments. Simultaneously as the elongation process happens, the DNA polymerase I goes back and replaces the RNA nucleotides with DNA nucleotides. Finally, a new enzyme called DNA ligase connects the Okazaki fragments together, making a continuous strand of DNA.
All the new nucleotides added are called deoxynucleioside triphosphates because they three phosphate group. When the 3’ end of the nucleotide is bonded to the triphosphate bonded to the 5’ end of the next nucleotide (because it runs in a 5’ to 3’ direction), two phosphates are dispensed to give energy for the bonding.
I would highly recommend watching a video on this as it is easier to understand in a visualized way. This is a very good video: https://www.youtube.com/watch?v=3jslVQDGkLU
DNA transcription
DNA encodes for proteins, but it is not DNA itself that is translated to proteins. It must first be transcribed to RNA through a process called transcription. A protein coding sequence in the DNA has a point where it starts and a point where it stops. To mark where to start transcribing, there is a sequence called the promoter. The DNA sequence that codes for the wanted protein lies on what is called the sense strand. But since DNA runs in antiparallel direction, it is the other strand (antisense strand) that has to be copied. If not, we would get an RNA sequence with the opposite code of what we wanted. The promoter always lies on the antisense strand, which is also known as the template strand. To mark the point where it should stop, there is a sequence called the terminator, which tells the transcription enzyme to stop. The enzyme that transcribes DNA into RNA is called RNA polymerase. RNA polymerase also runs in a 5’ to 3’ direction. The front of the enzyme unwinds the DNA, while the middle transcribes it onto RNA, and finally the back of the enzyme closes and rewinds the DNA.
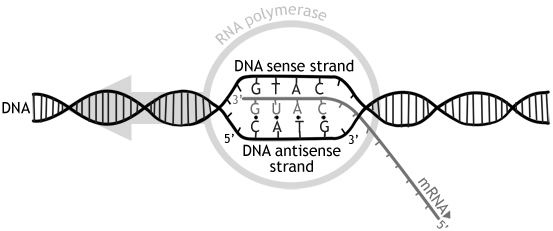
The resulting RNA sequence is called a mRNA (messenger RNA). However, it has to go through some processing before it can be translated to proteins. It is therefore called pre-mRNA at this point. The first step of processing also called modification, is to add a cap to the 5’ end, and a poly-A-tail to the 3’ end. The poly-A-tail is a sequence of around 50-250 adenine bases, and the cap and poly-A-tail seems to protect the mRNA from degradation. Additionally, as previously mentioned, DNA consists of coding and non-coding sequences. The coding sequences are called exons, while the non-coding sequences are called introns. This is a little ironic because the exons stay in, while the introns exit. Just remember that it is opposite of what is seems to be. The names do not come from in and out by the way, just so that is clear. Anyways, in a process called splicing, the introns are removed from the pre-mRNA by spliceosome enzymes composed of snRNA (snurps) and the exons are glued together to form mature mRNA. This mRNA then goes out of the nuclear envelope to be translated to proteins by ribosomes.
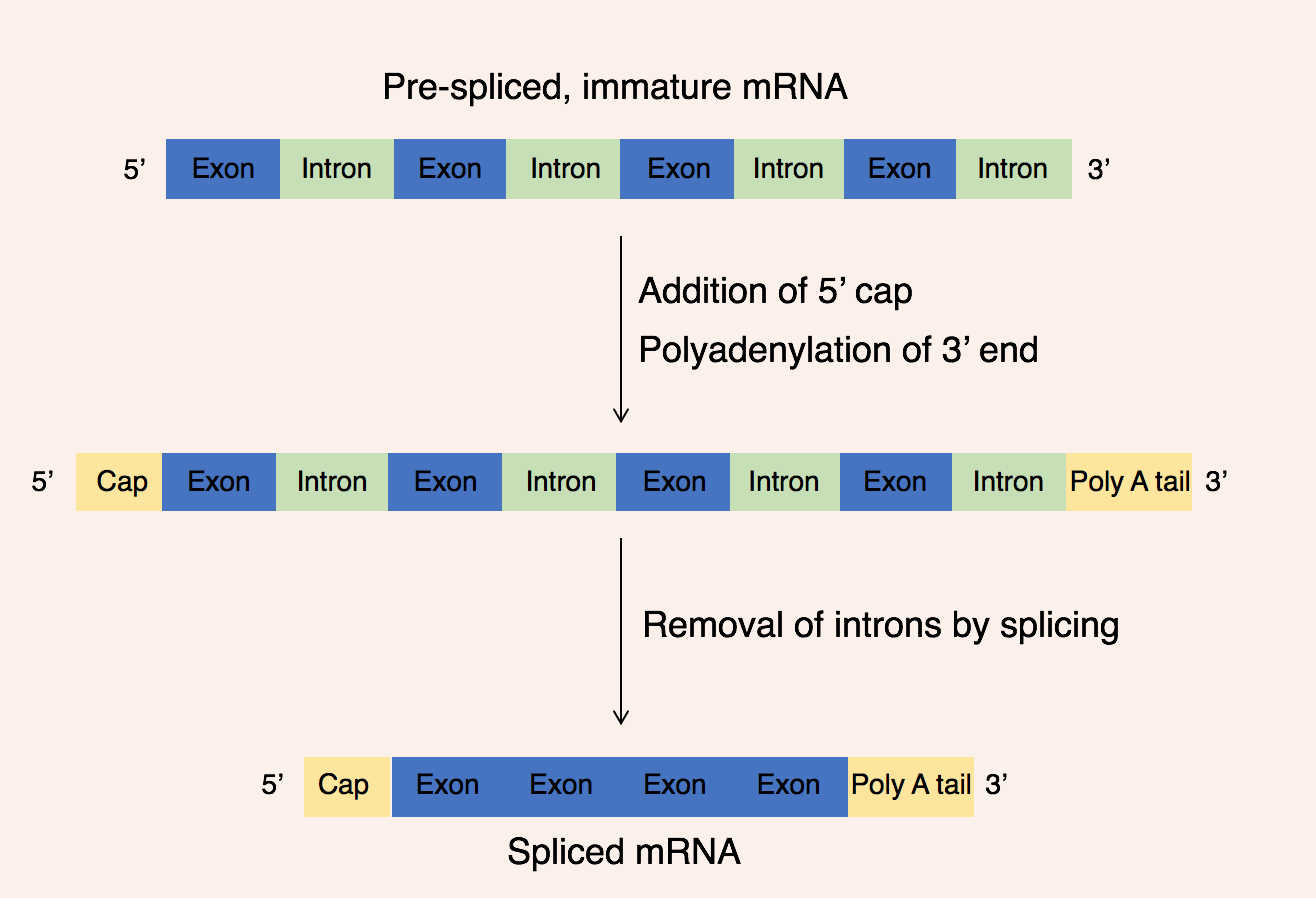
Translation and protein synthesis
Translation is the process that turns mRNA into proteins. As mentioned, pre-mRNA that has been processed into mature RNA is sent out of the nuclear membrane and attached to ribosomes. Ribosomes themselves are made of two subunits, conveniently named the large subunit and the small subunit. Both subunits are mainly comprised of rRNA (ribosomal RNA). The mRNA is squeezed between the large and small subunit, and it is in the cavity between where the polypeptide chain is assembled. The ribosome has three attachment sites called the A site, P site and E site. The mRNA strand enters the A site and exits the E site. mRNA is read in three and three base pairs called codons. Each codon codes for an amino acid with the exception of the start and stop codons, which signals where to start and stop.
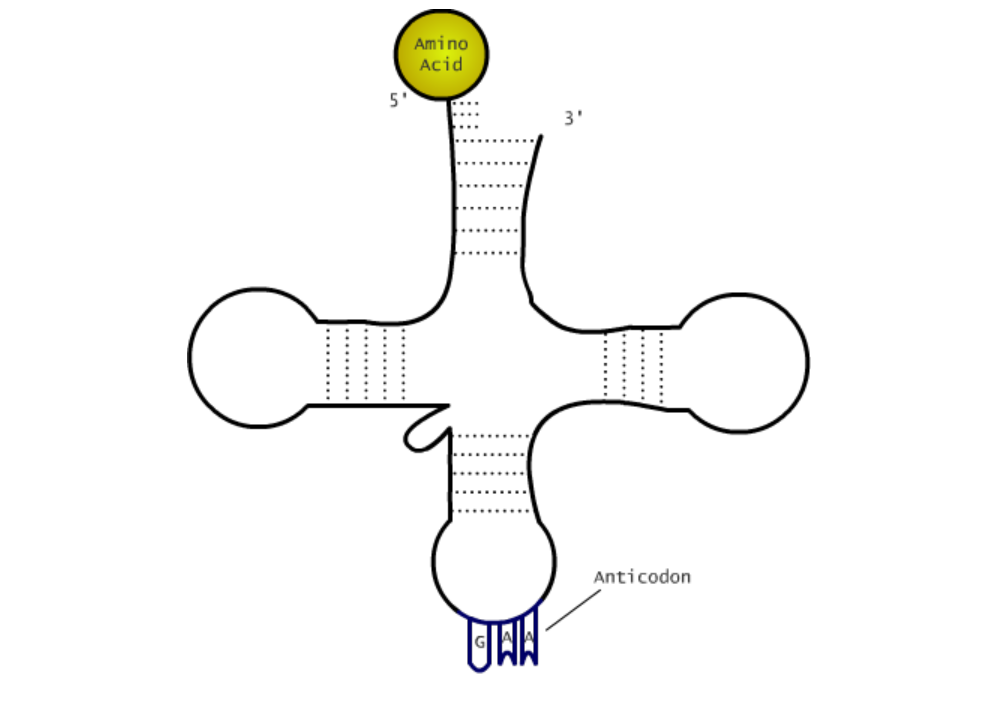
The first phase of translation is called the initiation phase, where the first amino acid is attached to the ribosome, thus starting the polypeptide chain. The start codon of an mRNA is always AUG, which codes for methionine. The start codon is on the 5’ end, which means that translation also goes in a 5’ to 3’ direction. Once the ribosome finds the start codon, a tRNA (transfer RNA) will bring the amino acid to the ribosome. tRNA has the shape of a shape of a clover, and the top of the clover has an anticodon which is specific to that tRNA. In the case of the start codon AUG, the anticodon has to be UAC. The tRNA with that anticodon will bring a methionine to the ribosome. The initiation complex is comprised of the ribosome, mRNA, initiation factors, and the first tRNA bonded to the mRNA by hydrogen bonds.
When the initiation phase is done, elongation and translocation occur. Elongation is the process of adding amino acids and is assisted by elongation factors that bind the tRNA to the mRNA. Translocation is the movement of tRNA through the ribosome. After the initiating tRNA is added, the ribosome moves to the next codon, which transfers the initiating tRNA to the P site, and adds a new tRNA with an amino acid to the A site. The amino acid on the initiating tRNA is then transferred to the new tRNA and the A site. When the ribosome moves to another codon, the initiating tRNA moves to the E site and is ejected. The amino acids in the polypeptide chain are bonded by peptide bonds. This process repeats until it reaches the last codon.
The last phase of translation is the termination phase, where one of the stop codons enter the A site. When this happens, release factors fill the A site, hydrolyzing the bond between the tRNA in the P site and the polypeptide chain. This releases the polypeptide chain and completes translation. The ribosomal complex is then deconstructed, and the mRNA and tRNA’s are released. A section about protein structure is covered in topic 2. Go back to revise if something is not clear.
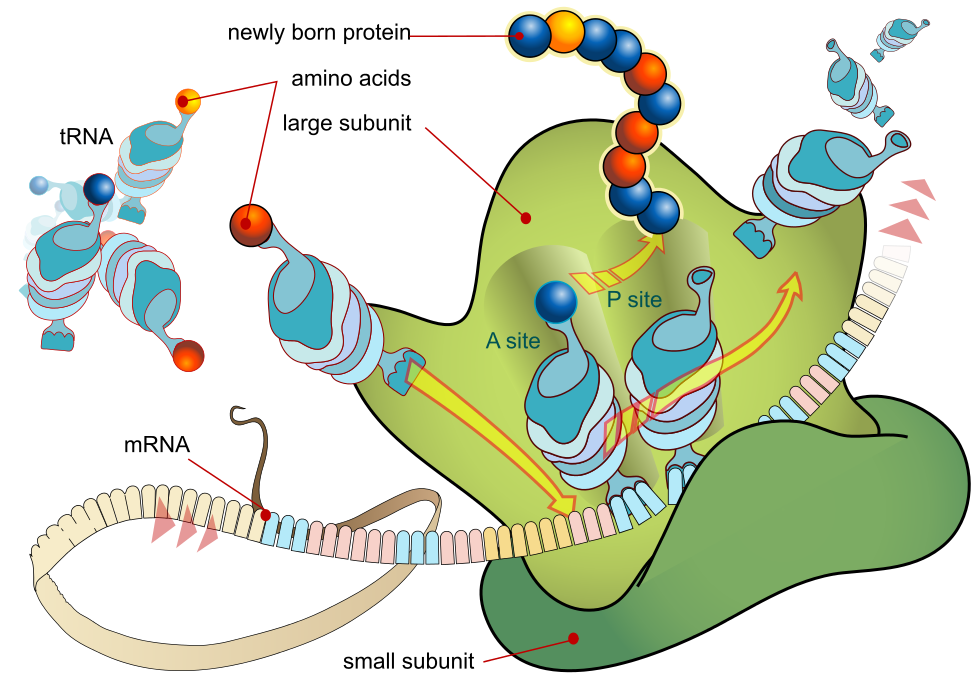
This is also a little hard to understand without visualization, so I would recommend watching this animation which shows it very well. https://www.youtube.com/watch?v=Ikq9AcBcohA
Excellent videos from MIT:
DNA replication: https://www.youtube.com/watch?v=DRBREvFL19g&list=LL&index=3
DNA transcription: https://www.youtube.com/watch?v=tMr9XH64rtM
Translation: https://www.youtube.com/watch?v=uBRdfsz_YB4
The diagrams and illustrations in this chapter are really the key to understanding the topic, as it takes some visualization to fully comprehend the information. I found it very useful to try to draw the structures and processes involved in transcription and translation, from DNA to protein. I would also HIGHLY recommend the MIT videos I linked to, as Professor Lander does an excellent job at explaining the topics. Otherwise, is way a challenging chapter, but I liked it very much.